Multiscale protein-protein interactions
Disorder & aggregation
CSULA Seminar: February 3, 2015
National Institutes of Health (NIH)
National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK)
Laboratory of Chemical Physics (LCP), Theoretical Biophysical Chemistry (TBC)
Biophysical question #1
How do we predict phase separations of protein solutions?
Biophysical question #2
How do we make predictions about intrinsically disordered proteins
given their large conformational landscape?
Acknowledgments:
Laboratory of Chemical Physics


Laboratory of Biochemistry and Genetics


*Support provided by the Intramural Research Division of the NIDDK, NIH.
Protein Structure
Primary structure (sequence)
GSIGAASMEF CFDVFKELKV HHANENIFYC PIAIMSALAM VYLGAKDSTR TQINKVVRFD KLPGFGDEIE AQCGTSVNVH
SSLRDILNQI TKPNDVYSFS LASRLYAEER YPILPEYLQC VKELYRGGLE PINFQTAADQ ARELINSWVE SQTNGIIRNV
LQPSSVDSQT AMVLVNAIVF KGLWEKAFKD EDTQAMPFRV TEQESKPVQM MYQIGLFRVA SMASEKMKIL ELPFASGTMS
MLVLLPDEVS GLEQLESIIN FEKLTEWTSS NVMEERKIKV YLPRMKMEEK YNLTSVLMAM GITDVFSSSA NLSGISSAES
LKISQAVHAA HAEINEAGRE VVGGAEAGVD AASVSEEFRA DHPFLFCIKH IATNAVLFFG RCVSP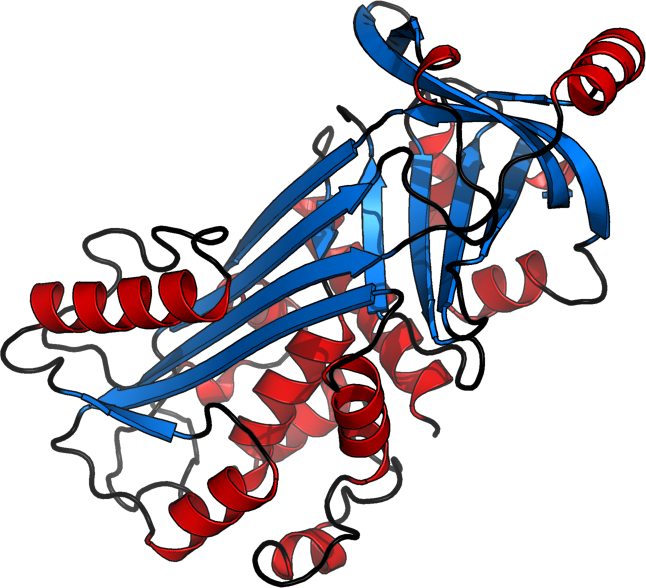
helices [red], sheets [blue
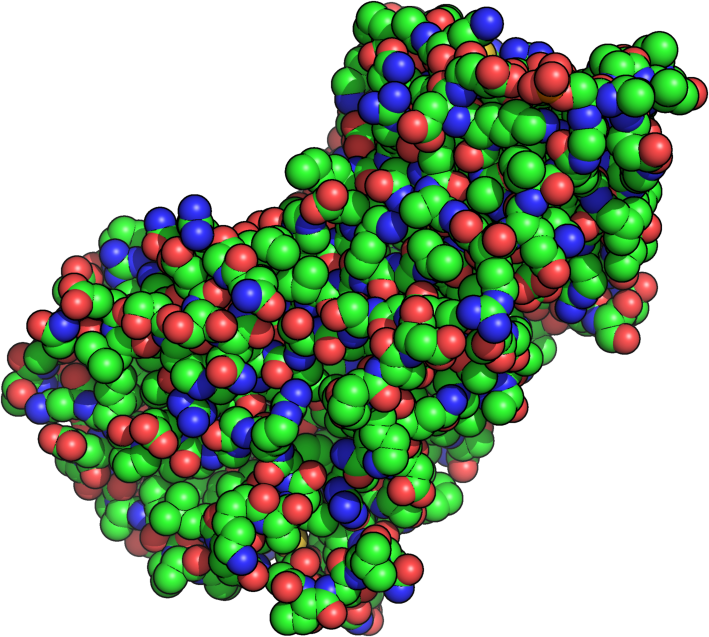
3D structure
complexes, aggregation
SEM Aggregate structure, Zabik et al., J. Poul. Sci. (1980)
Primary Structure
Twenty residue "alphabet" forms polypeptide chain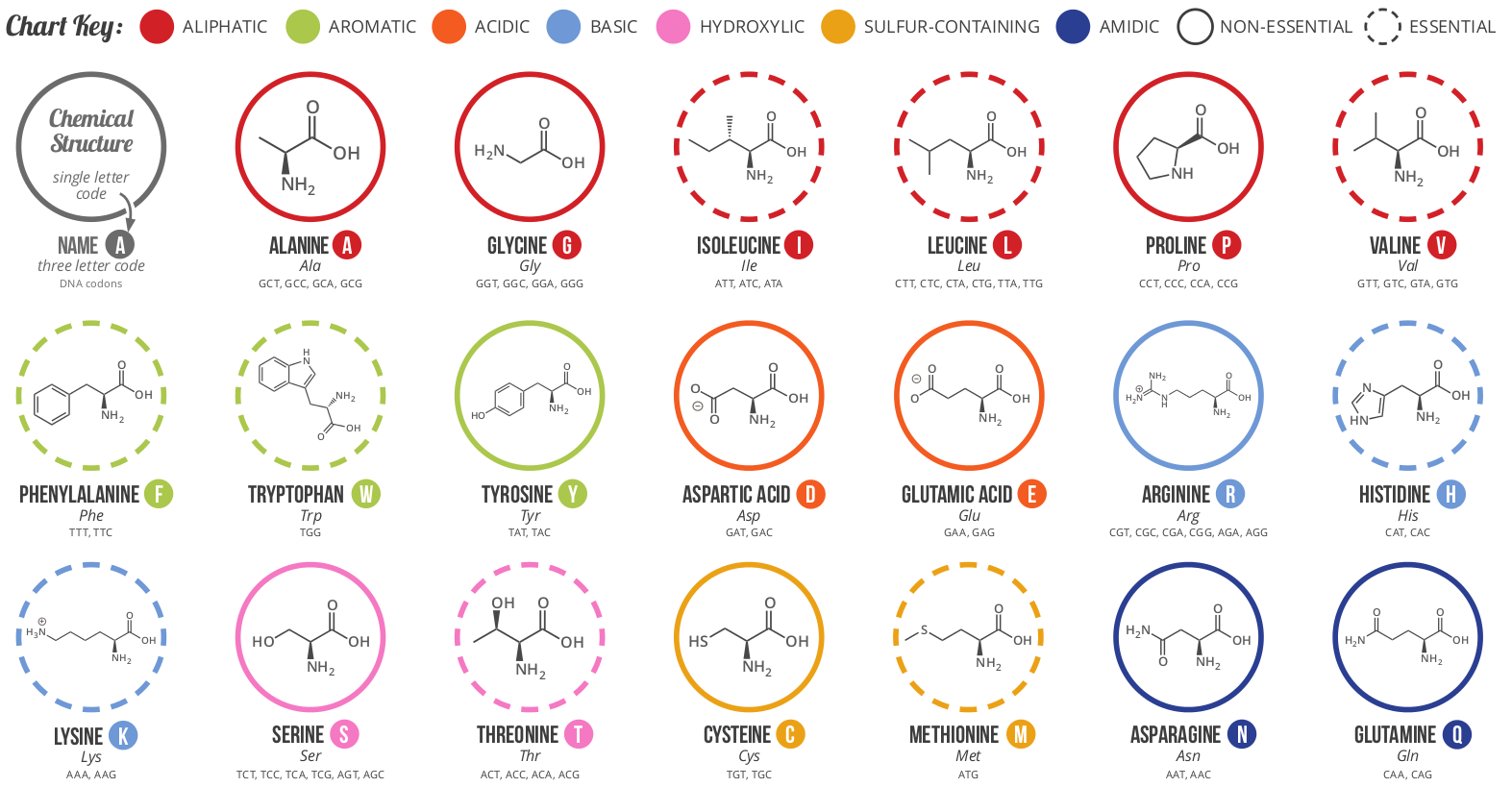
Protein folding problem
Predict structure from sequence
Sequence Structure Function
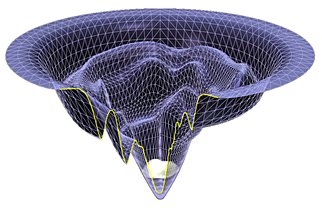
Native structure, folding pathways, ...
MD simulation of WW-domain, Best and Mittal, J. Phys. Chem. B (2010)
Scientific Philosophy
Theoreticians need to keep in close contact with experimentalists.Imagination must be constrained by reality.
Part 1: Aggregation
How do we predict phase separations of protein solutions?
Higher order structure
Phase separations lead to sudden changes in liquid structure.

How do we model many protein-protein interactions?
Can we predict aggregates from experimental structure?
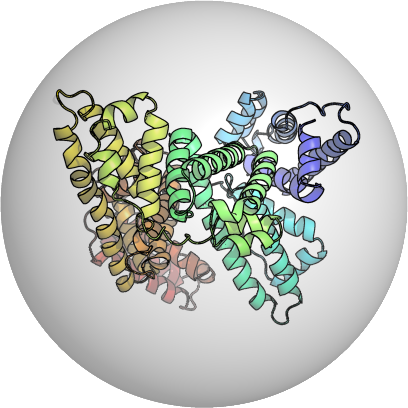
PDB:1AO6
PDB:1OVA

PDB:1W6Z
PDB:3V03
Protein-Protein interactions
Important terms:
Non-specific interactions (London/dispersion forces)
Second-order effects?
internal conformational energies, ...
Need a way of validating model.
Experimental Measurements
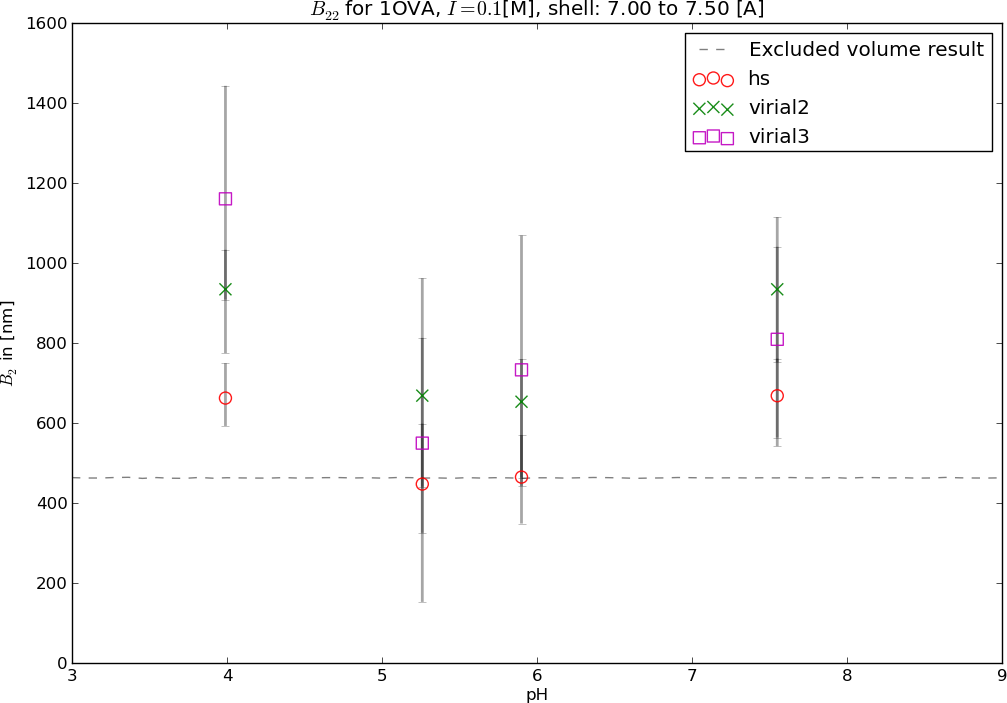
Second virial coefficient, , measurementusing light scattering at different pH.
Virial Coefficients
An equation of state expanded in powers of density
is the pairwise interaction of two moleculesis the interaction of three molecules, ...
Negative values of often correlate with aggregation.
For rotationally invariant molecules*
Goal: Develop a realistic pair potential for virial calculation.
The Process

Start with the crystallized PDB Structure
e.g. Human Serum Albumin PDB:1A06
Electrostatics: Poisson-Boltzmann
Solve for with the Adaptive Poisson-Boltzmann Solver (APBS),
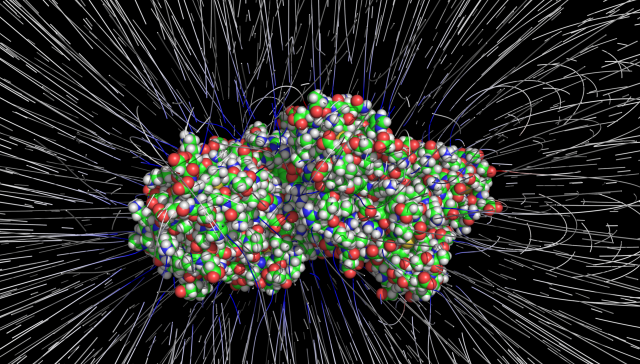
Typically (in the absence of ions), and .
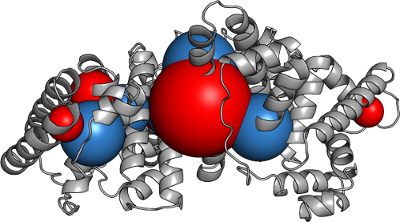
Macrocharge fitting
Best fit macrocharges to approximate the field.
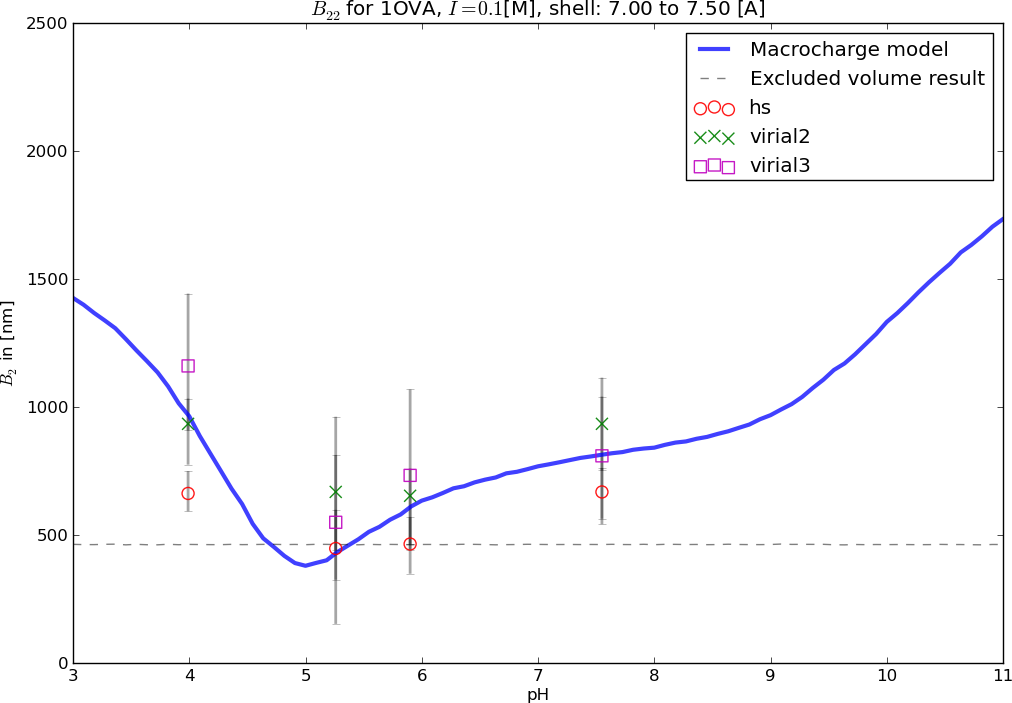
Matching experiments
Theoretical predictions of the second virial coefficientconsidering only excluded volume and reduced electrostatics.

Matching experiments
Theoretical predictions of the second virial coefficientconsidering only excluded volume and reduced electrostatics.
Phase separations summary
Calculate the non-ideality of a protein molecule after including
both the excluded volume and electrostatics.
Predict the second-virial coefficient as a function of pH values, protein concentrations, binary mixtures, and salt concentrations.
Ongoing research: Use the model in higher-order simulations
to predict phase behavior via Gibb's ensembles.
Part 2: Disorder
How do we make predictions about intrinsically disordered proteins
given their large conformational landscape?
Paradigm shift
Proteins were thought to adopt stable, folded conformations.Solving the structure was paramount for understanding the function.
Intrinsically disordered proteins
Structure
- Lack tertiary structure (disorder!)
- Still may form secondary structure
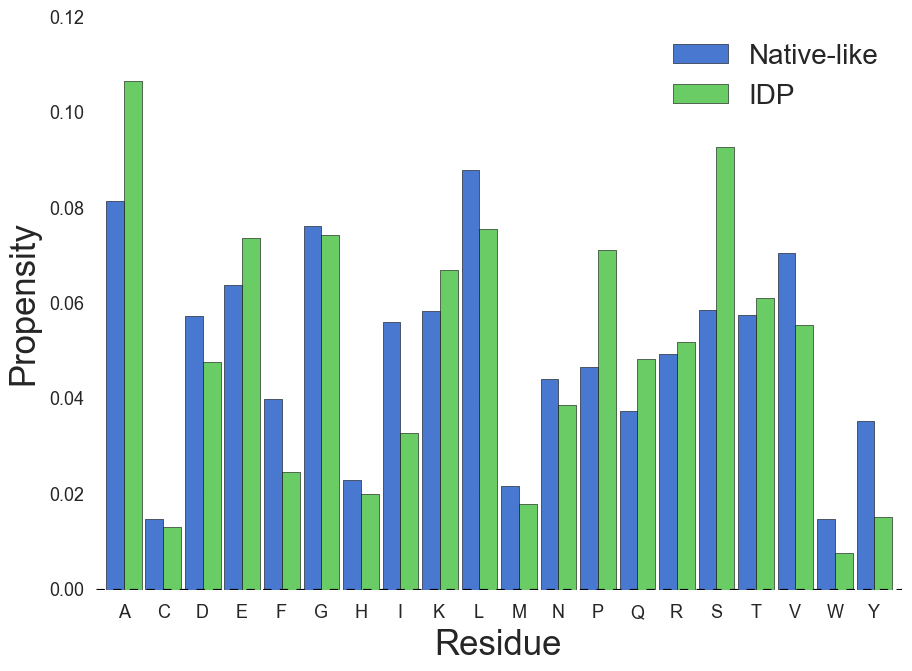
- Different primary structure (residue propensity)
- More charged, less hydrophobic and aromatic residues
Binding
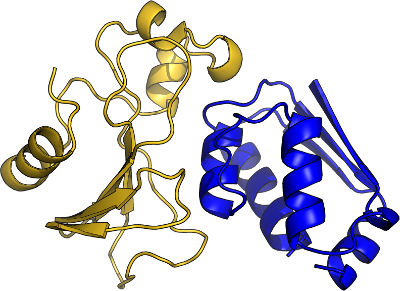
Barnase-Barstar complex
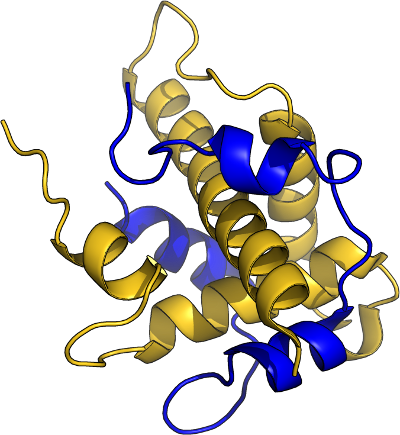
Hif-1 α/CBP
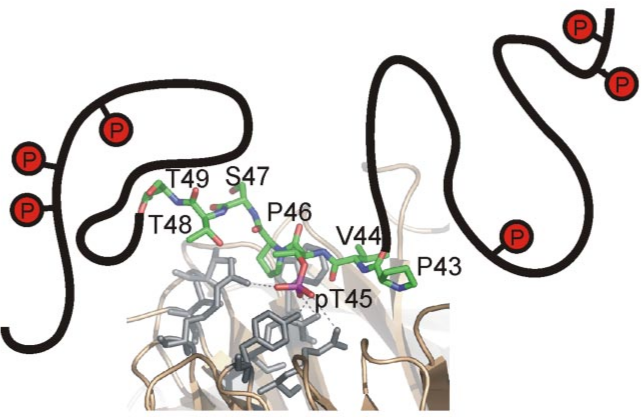
SIC1 binding to CDC4
Theory
- What advantages do IDPs have over traditional proteins?
- Recognition that the cellular environment is a crowded place.
Function
- Often found in signaling pathways, centers of protein hubs
- Linkers (entropic chains), Chaperones, HIV transcription (TAT)
- Binding specificity, with lower affinity
Modeling
IDPs: Folding Sampling
Goal: Develop a model for IDP interactions.
Statistical Potentials
Residue-residue interactions, quasi-chemical lattice-gas
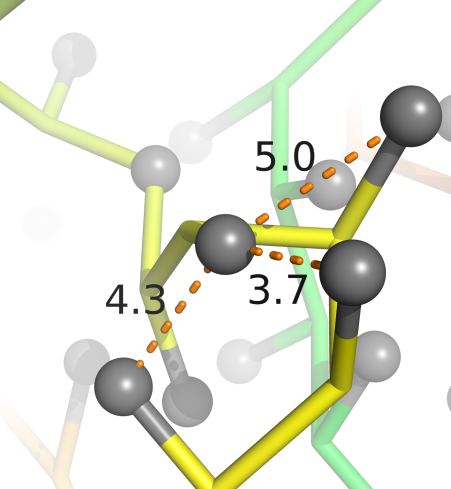
Residue-residue interaction matrix, MJ
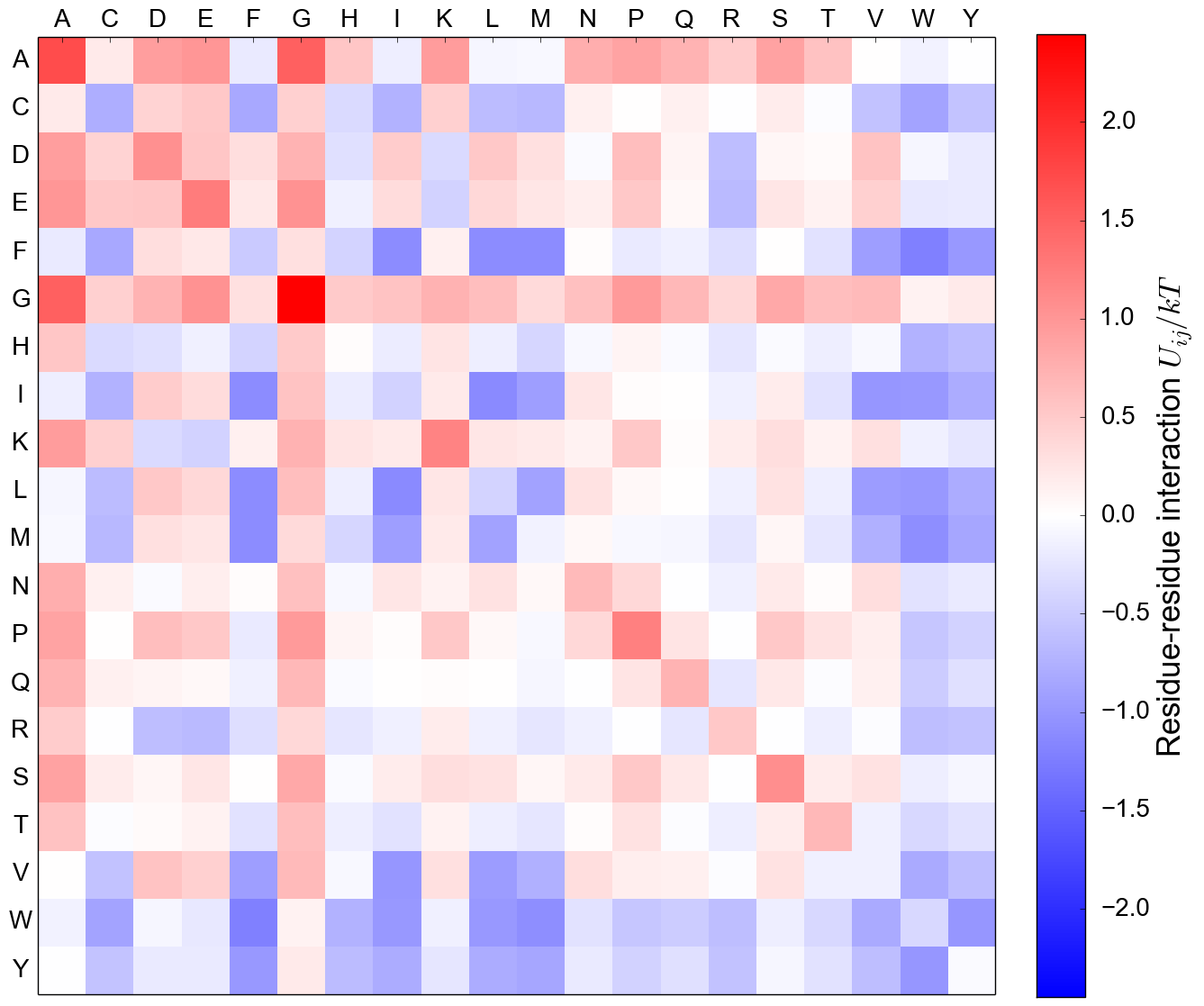
Betancourt and Thirumalai (1999), Skolnick, Kolinski and Ortiz (2000)
MJ matrix reveals biophysical structure
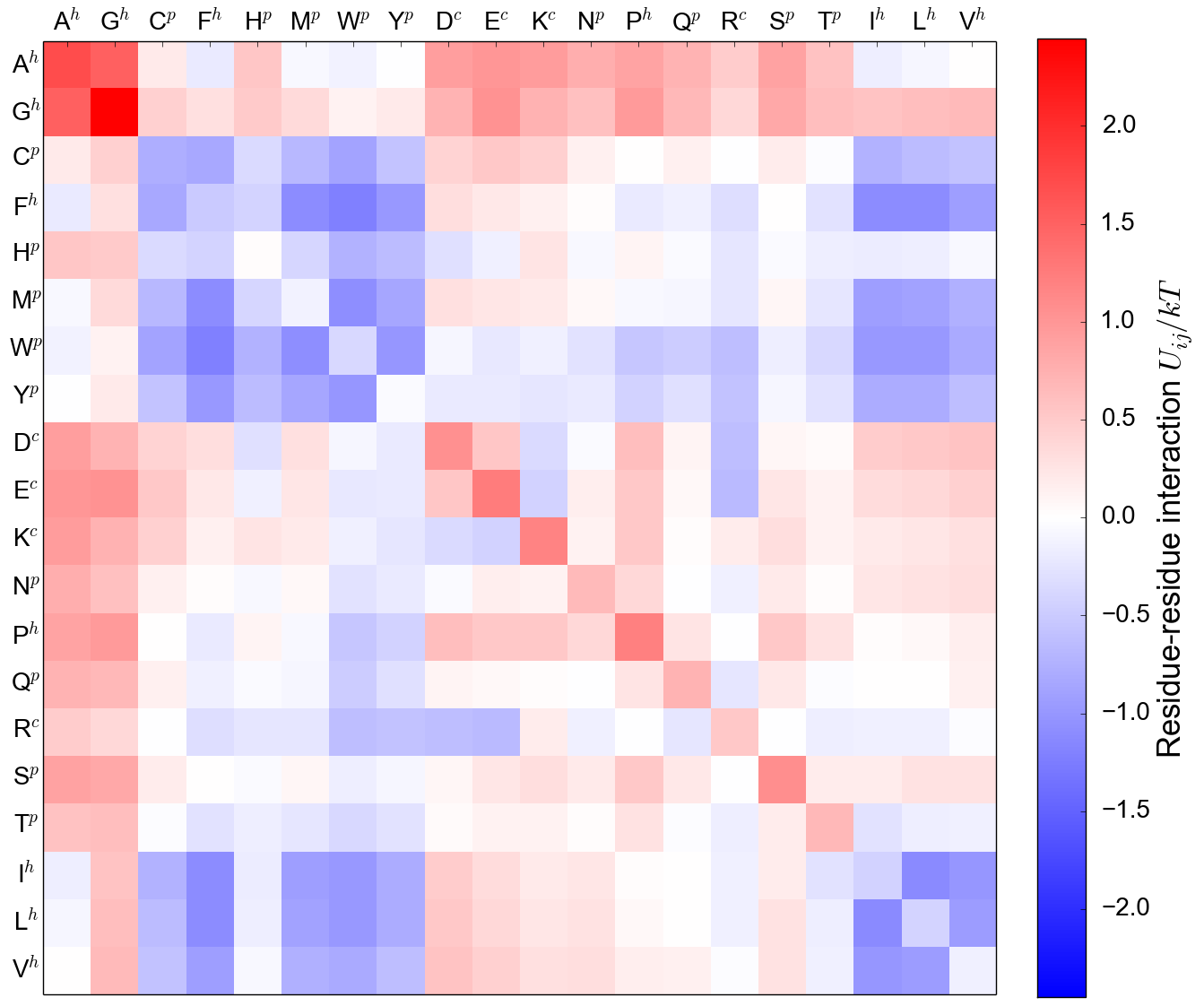
MJ Contact energy, from structure
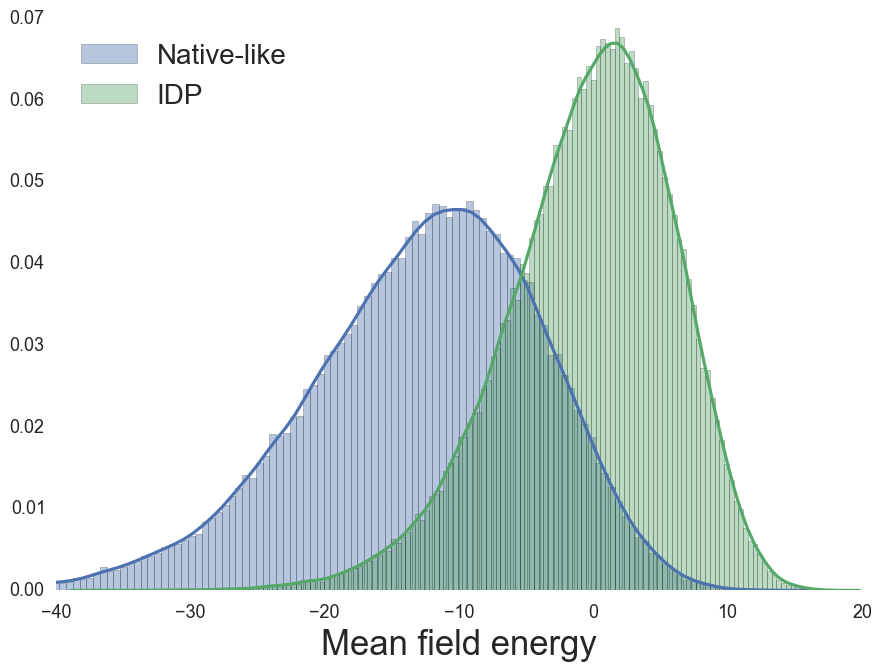
Mean-field (MF) energy, from sequence
MJ contact energy reproduces MF energy
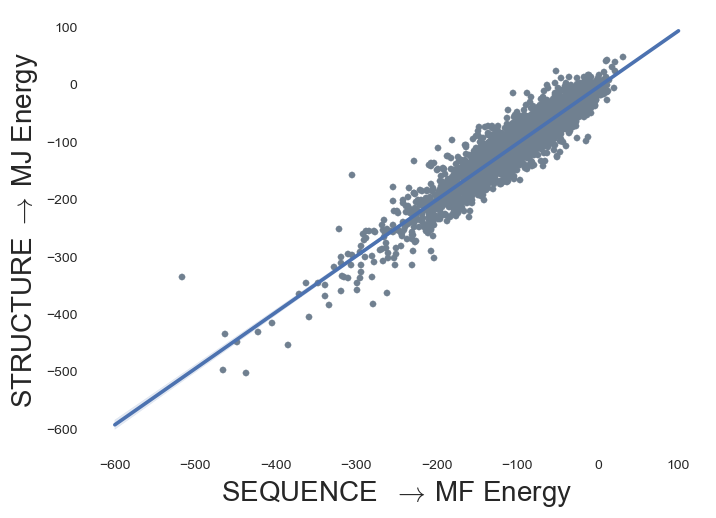
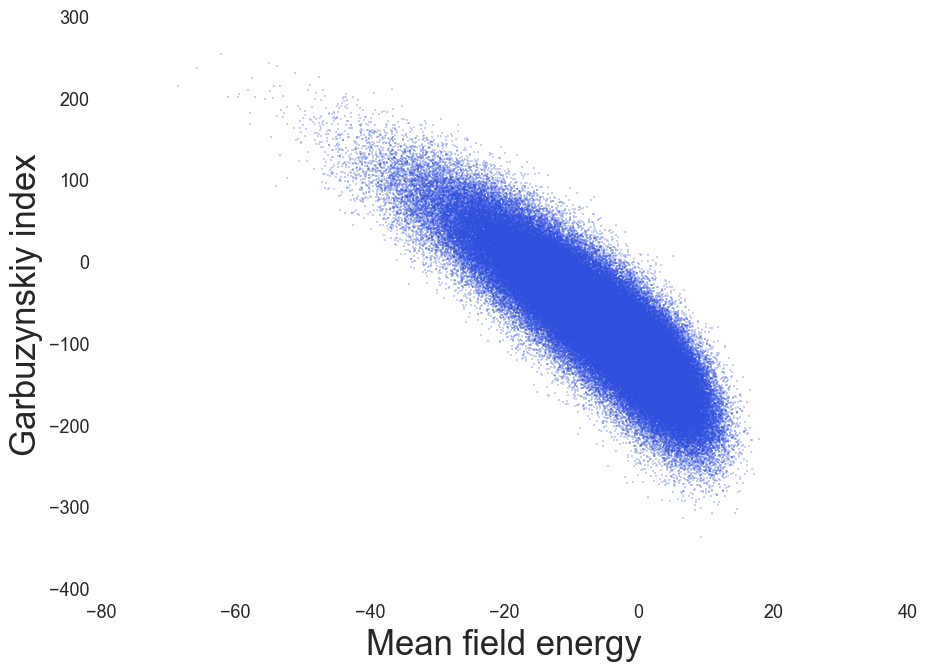
MF Energy distributions: Physically reasonable
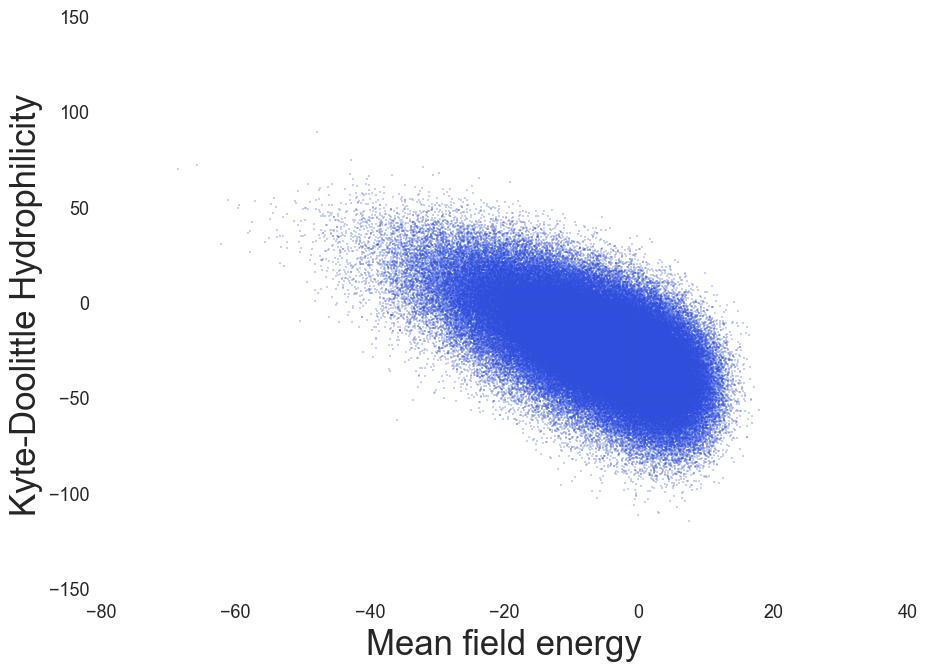
Hydrophilicity index, Kyte & Doolittle, J. Mol. Biol. (1982)
Amyloidogenic regions, Garbuzynskiy et. al. Bioinformatics (2010)
Protein Networks
- Target protein interacts with a range of possible surfaces.
- Measure average binding affinity of protein to surfaces.
- Measure binding specificity of protein to surfaces.
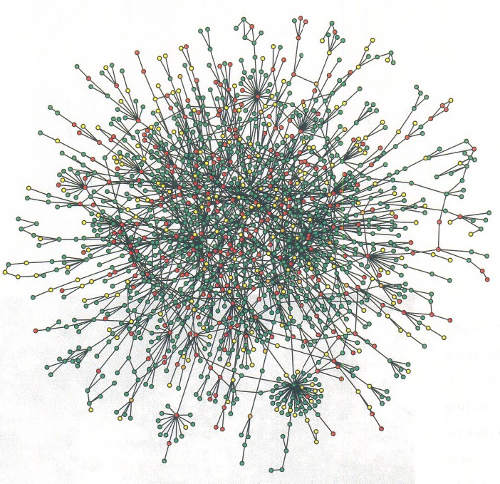
Schwikowski & Fields et al., Nature 2000.
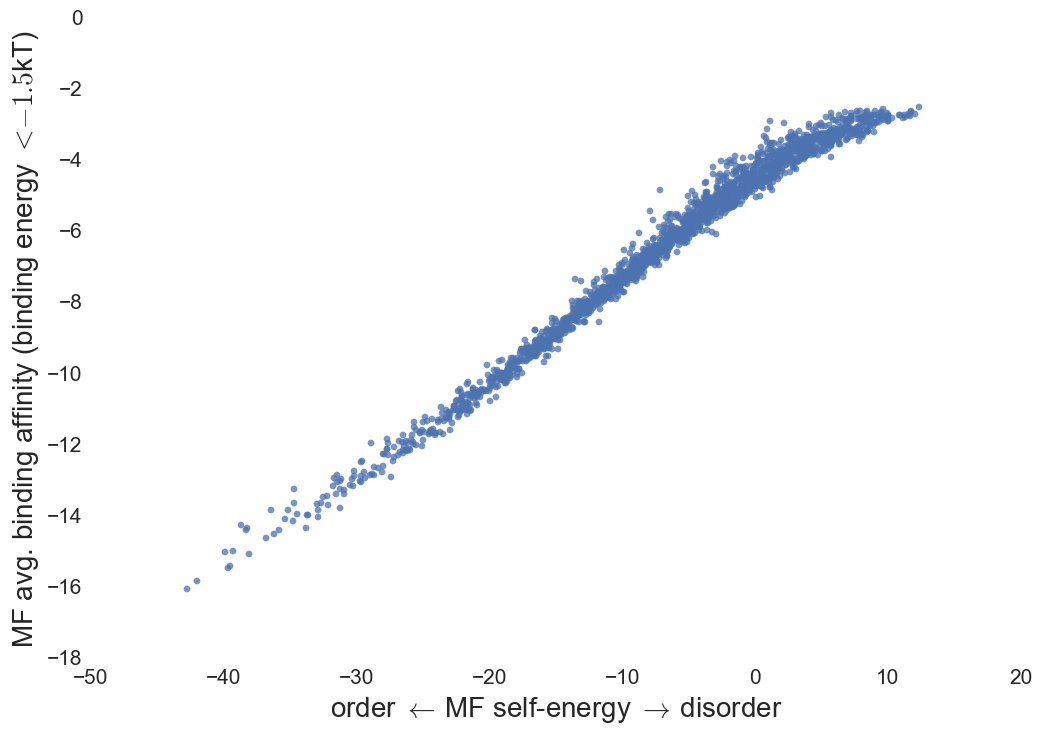
Protein-complex energy
Pairwise decomposition of protein complex energy; Binding affinity
Contact matrix is not symmetric
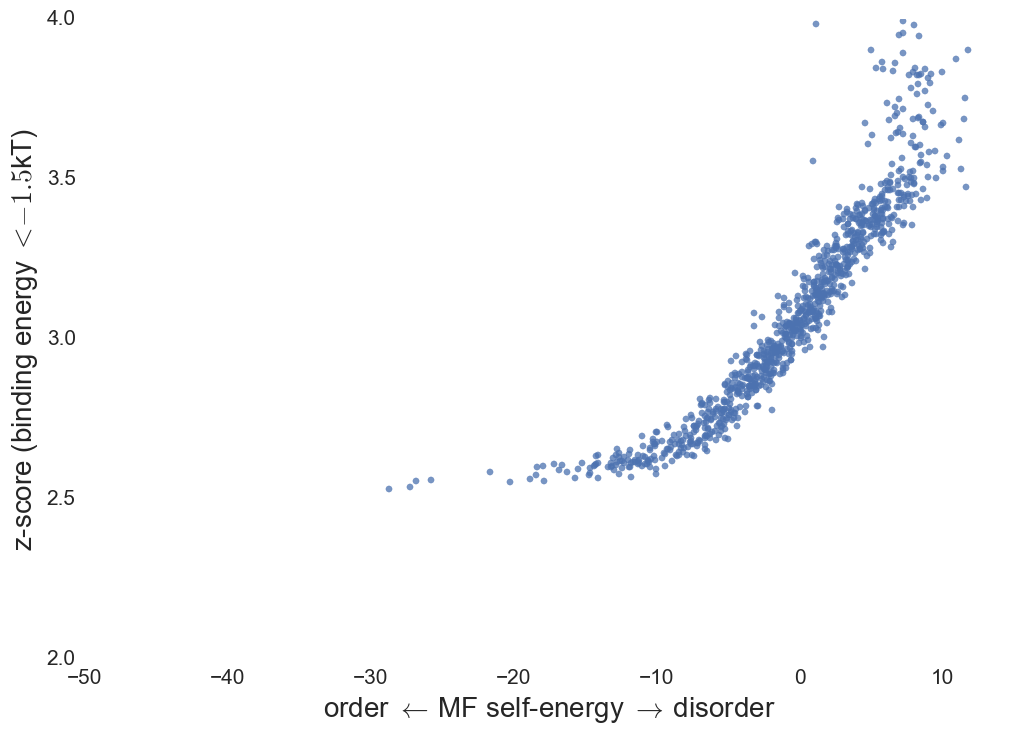
Specificity score: Define "decoys" as weakly bound
structures in protein network.
Binding affinity
Binding specificity
MF IDP Summary:
- MF models reproduce MJ contact energies. MF IDP's bound to native structures show increased specificity with lower affinity.
Active research projects & collaborations
Crowding, surface adsorption and protein fibrillation, Biophys (in press).Programmable Nanoscaffolds and Multivalent Effects, JACS.
Integer sequence discovery from small graphs, Discrete Math. (submitted).
Dependence of Internal Friction on Folding Mechanism, JACS (submitted).
Quantification of plasma HIV RNA, Nature Comm.
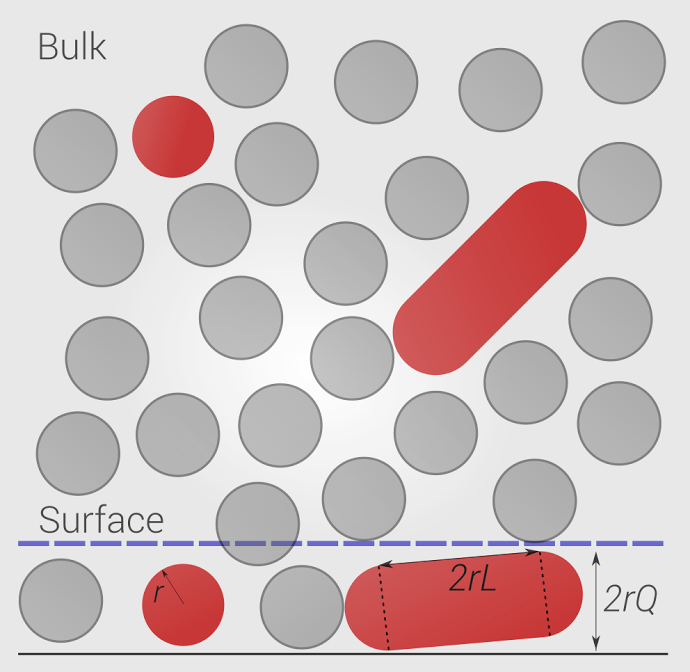
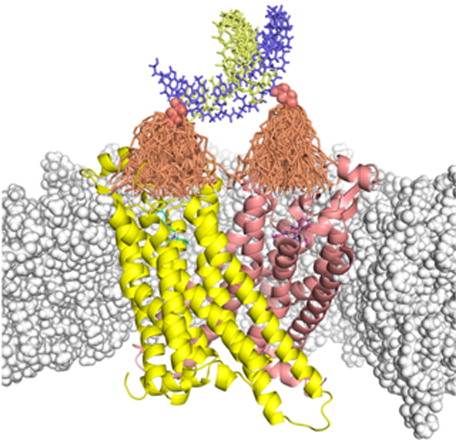
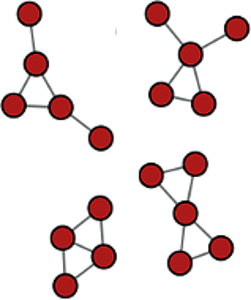
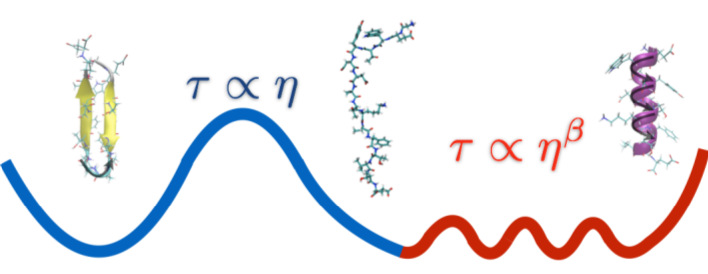
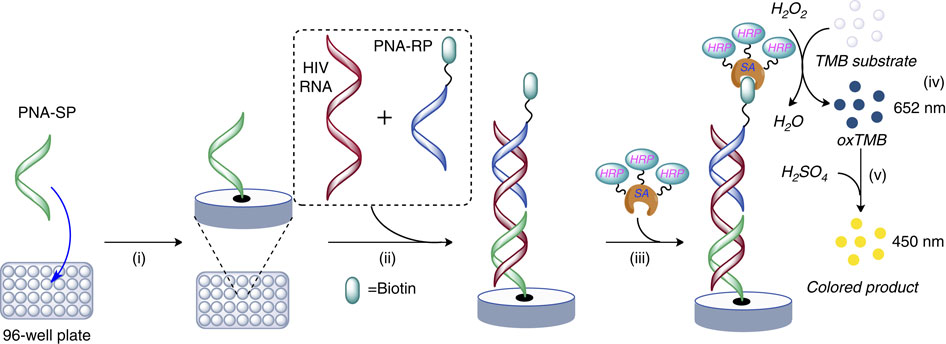
Future Research Projects
Phase separation calculations, aggregation.
Quantitative IDP models, disorder.
Benchmarks in sampling algorithms, BiSA.
Graph fingerprint and invariant database, EoFG.
Dependence of topology on sampling, WL topology.
RNA structure as multigraphs.
Theoretical liquid state calculations for simple potentials.
Entropic microscopes: free chain calculations of PNA.