Mean-field lattice-model IDPs
Binding Affinity & Specificity
Travis Hoppe, Robert Best
Biophysical Society Meeting, 2015
National Institutes of Health (NIH)
National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK)
Laboratory of Chemical Physics (LCP), Theoretical Biophysical Chemistry (TBC)
Slide source at https://github.com/thoppe/Presentation_Research_IDP
Intrinsically disordered proteins
Structure
- Lack tertiary structure (disorder!)
- Different primary structure (residue propensity)
- More charged, less hydrophobic and aromatic residues
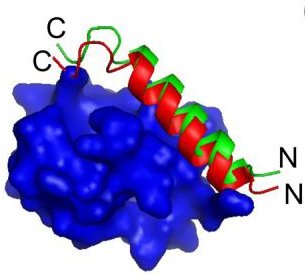
Binding
Function
- Linkers (entropic chains), Chaperones, HIV transcription (TAT)
- Often found in signaling pathways, centers of protein hubs
- Binding specificity, with lower affinity
Questions
- What advantages do IDPs have over traditional proteins?
- How to incorporate the crowded cellular environment?
Modeling
IDPs: Folding Sampling
Goal: Develop a model for IDP binding interactions.
Statistical Potentials
Residue-residue interactions, quasi-chemical lattice-gas
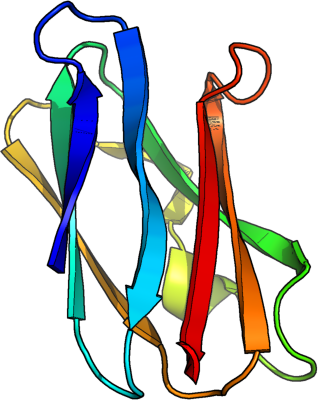
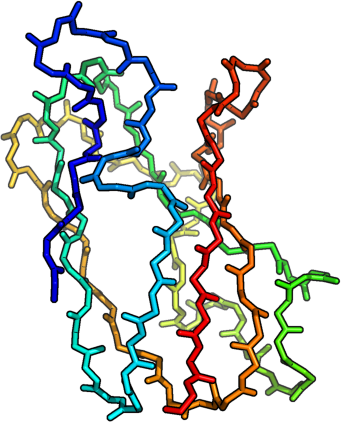
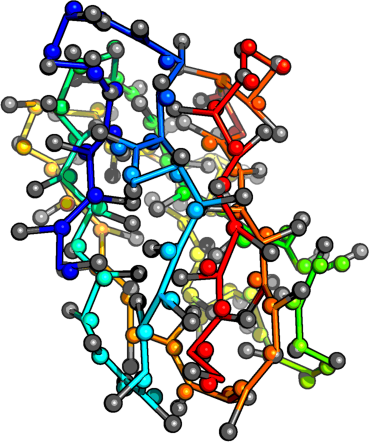
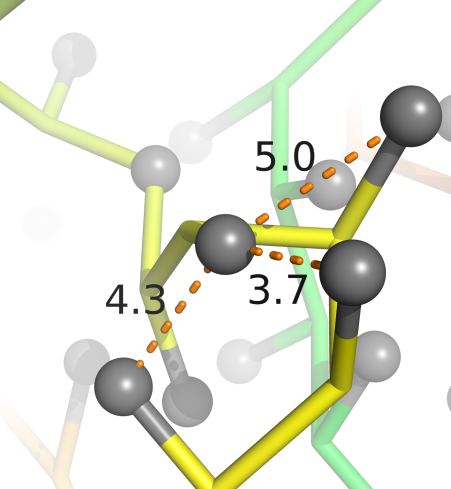
Potentials constructed from Top 8000 Protein Database, Richardson Group
Statistical potentials have predictive power
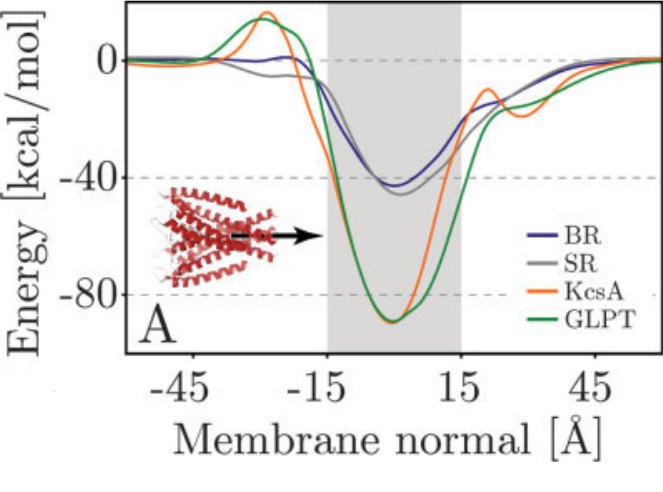
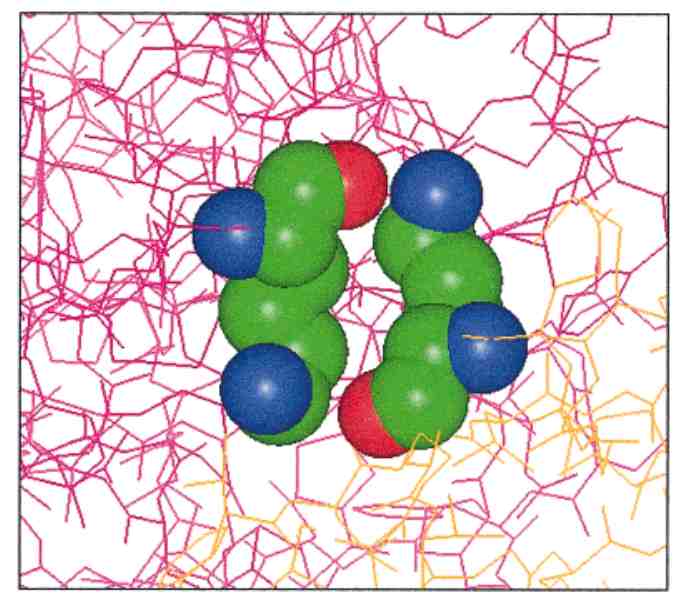
[1] Coarse-grained models for simulations of multiprotein complexes, Kim & Hummer, J Mol Biol(375):1416-33
[2] Properties of Integral Membrane Protein Structures, Ulmschneider, Sansom & Nola, Proteins(59):252-265
[3] Pairing preferences at protein-protein interfaces, Glaser, et al., Proteins:43(2) 89-102
[2] Properties of Integral Membrane Protein Structures, Ulmschneider, Sansom & Nola, Proteins(59):252-265
[3] Pairing preferences at protein-protein interfaces, Glaser, et al., Proteins:43(2) 89-102
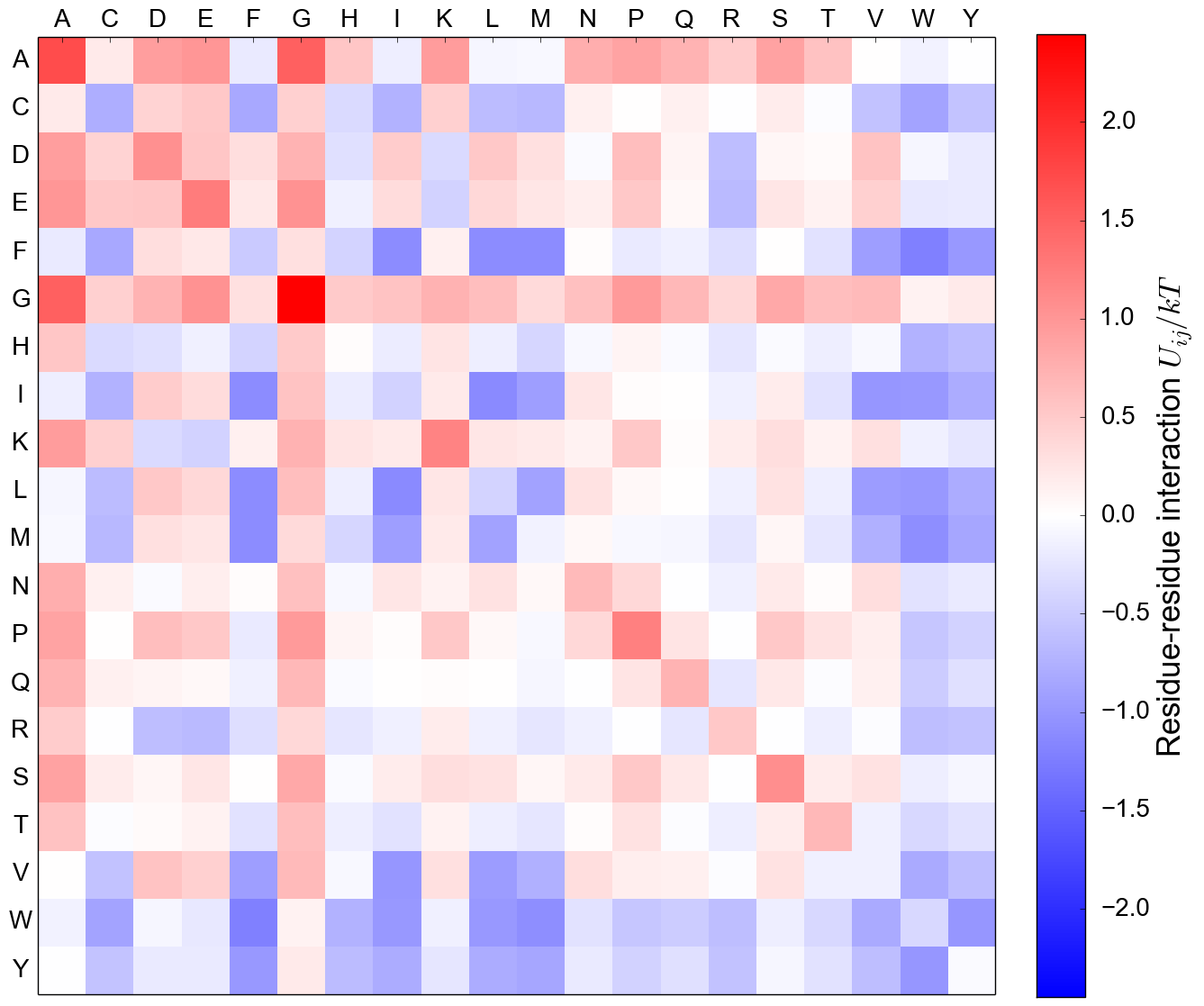
Residue-residue interaction matrix, MJ
Other statistical potentials: Tanaka and Scheraga (1976), Spil (1990), Miyazawa and Jernigan (1996),
Betancourt and Thirumalai (1999), Skolnick, Kolinski and Ortiz (2000)
Betancourt and Thirumalai (1999), Skolnick, Kolinski and Ortiz (2000)
MJ Contact energy, from structure
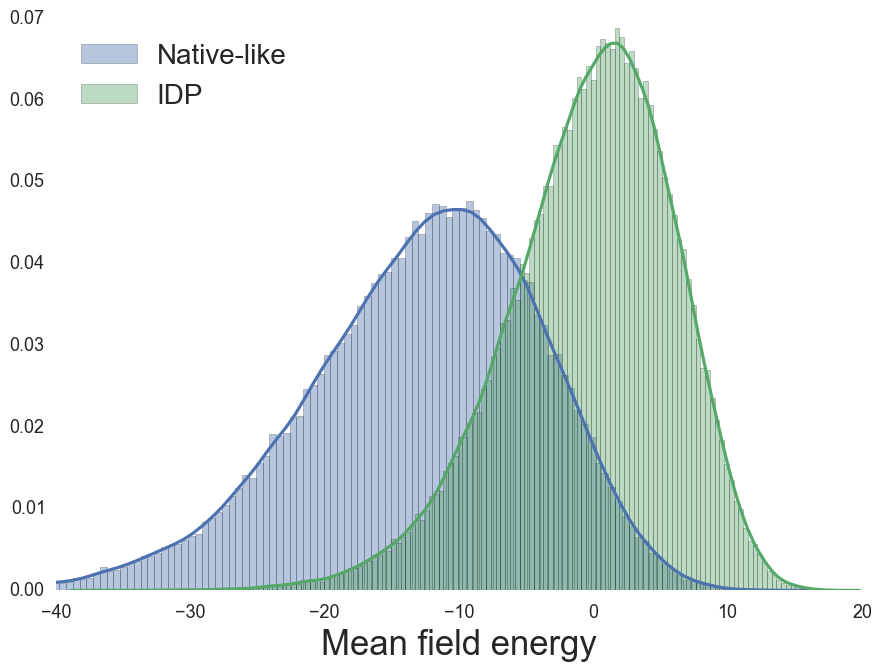
Mean-field (MF) energy, from sequence
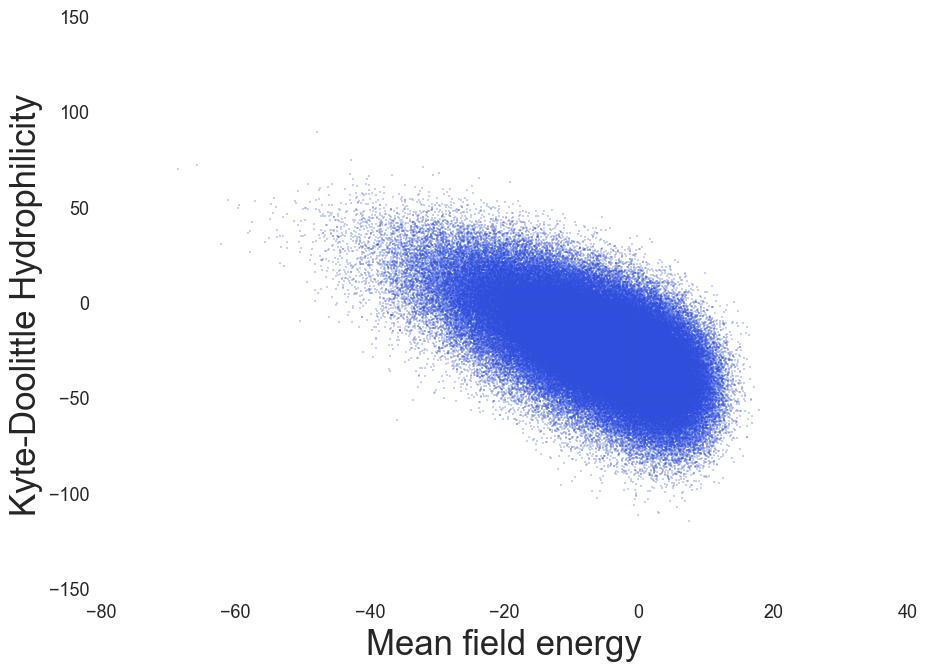
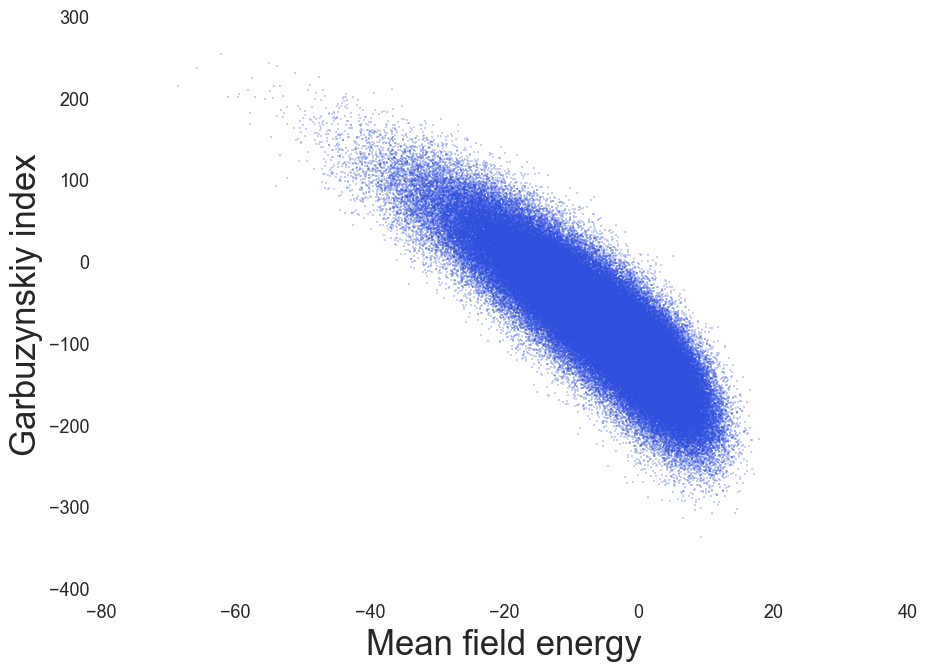
MF Energy distributions: Physically reasonable
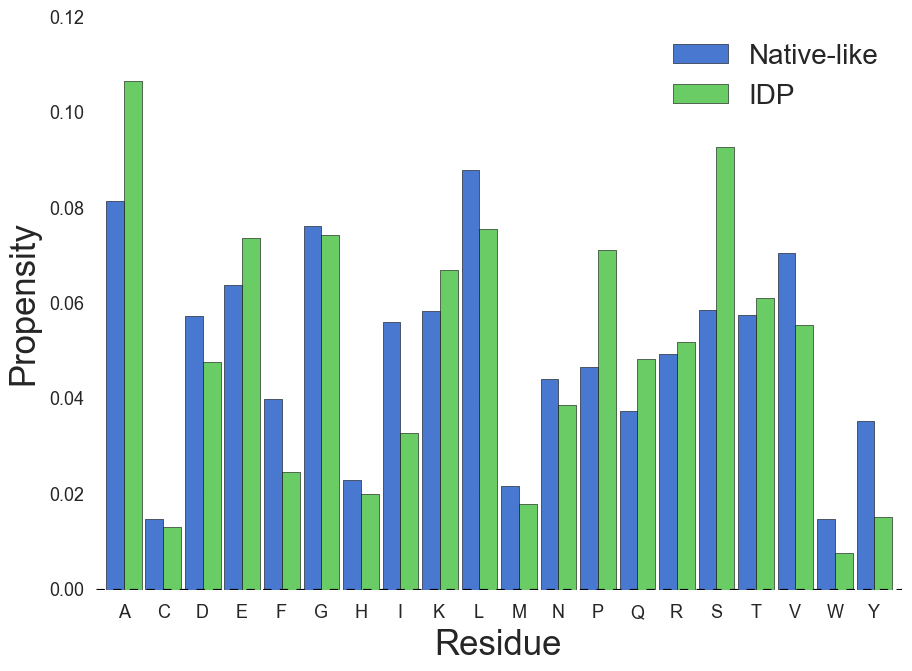
IDP Propensity, Coeytaux & Poupon, Bioinformatics (2005)
Hydrophilicity index, Kyte & Doolittle, J. Mol. Biol. (1982)
Amyloidogenic regions, Garbuzynskiy et. al. Bioinformatics (2010)
Hydrophilicity index, Kyte & Doolittle, J. Mol. Biol. (1982)
Amyloidogenic regions, Garbuzynskiy et. al. Bioinformatics (2010)
Protein Networks
- Target protein interacts with a range of possible surfaces.
- Measure average binding affinity of protein to surfaces.
- Measure binding specificity of protein to surfaces.
- A priori structure is unknown; simplest model first.
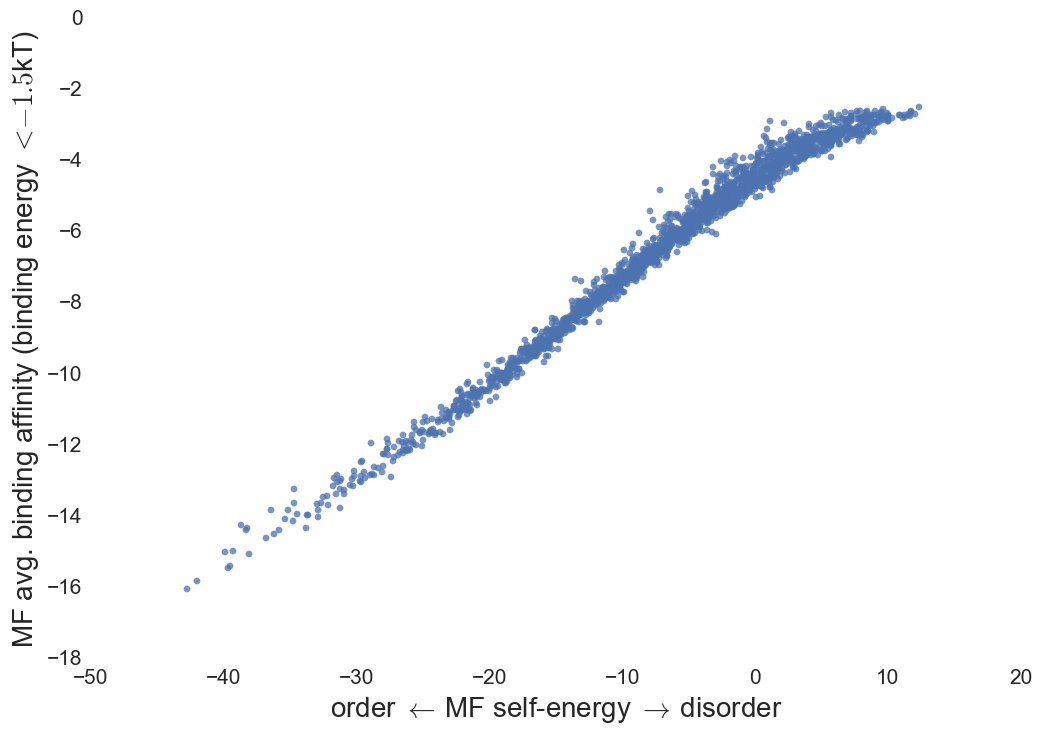
Protein-complex energy
Pairwise decomposition of protein complex energy; Binding affinity
Contact matrix is not symmetric
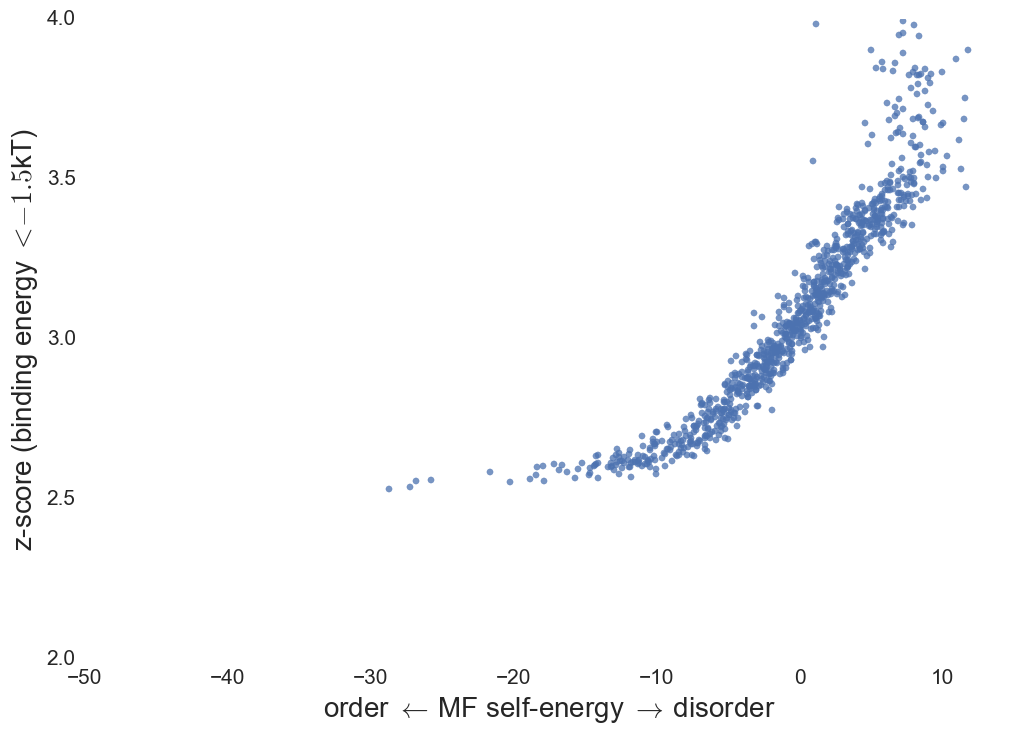
Specificity score: Define "decoys" as weakly bound
structures in protein network.
Binding affinity
Binding specificity
Summary & Future Work
- MF IDP's bound to native structures show increased
specificity with lower affinity.
Thank you.
Questions?Laboratory of Chemical Physics